- Acne
- Actinic Keratosis
- Aesthetics
- Alopecia
- Atopic Dermatitis
- Buy-and-Bill
- COVID-19
- Case-Based Roundtable
- Chronic Hand Eczema
- Drug Watch
- Eczema
- General Dermatology
- Hidradenitis Suppurativa
- Melasma
- NP and PA
- Pediatric Dermatology
- Pigmentary Disorders
- Practice Management
- Precision Medicine and Biologics
- Prurigo Nodularis
- Psoriasis
- Psoriatic Arthritis
- Rare Disease
- Rosacea
- Skin Cancer
- Vitiligo
- Wound Care
News
Article
Deep Learning Tool Differentiates Keloids From Malignant Lesions Without Dermoscopy
Author(s):
Key Takeaways
- The VGG16 CNN model showed high accuracy in distinguishing keloids from other skin lesions, achieving an AUC of 0.996 and precision of 1.0.
- Challenges include image quality and limited data, which may affect the model's generalizability and performance.
After fine-tuning, the VGG16 convolutional neural network outperformed other deep learning models in 2 of 3 key classification tasks.
Image credit: weerajata – stock.adobe.com
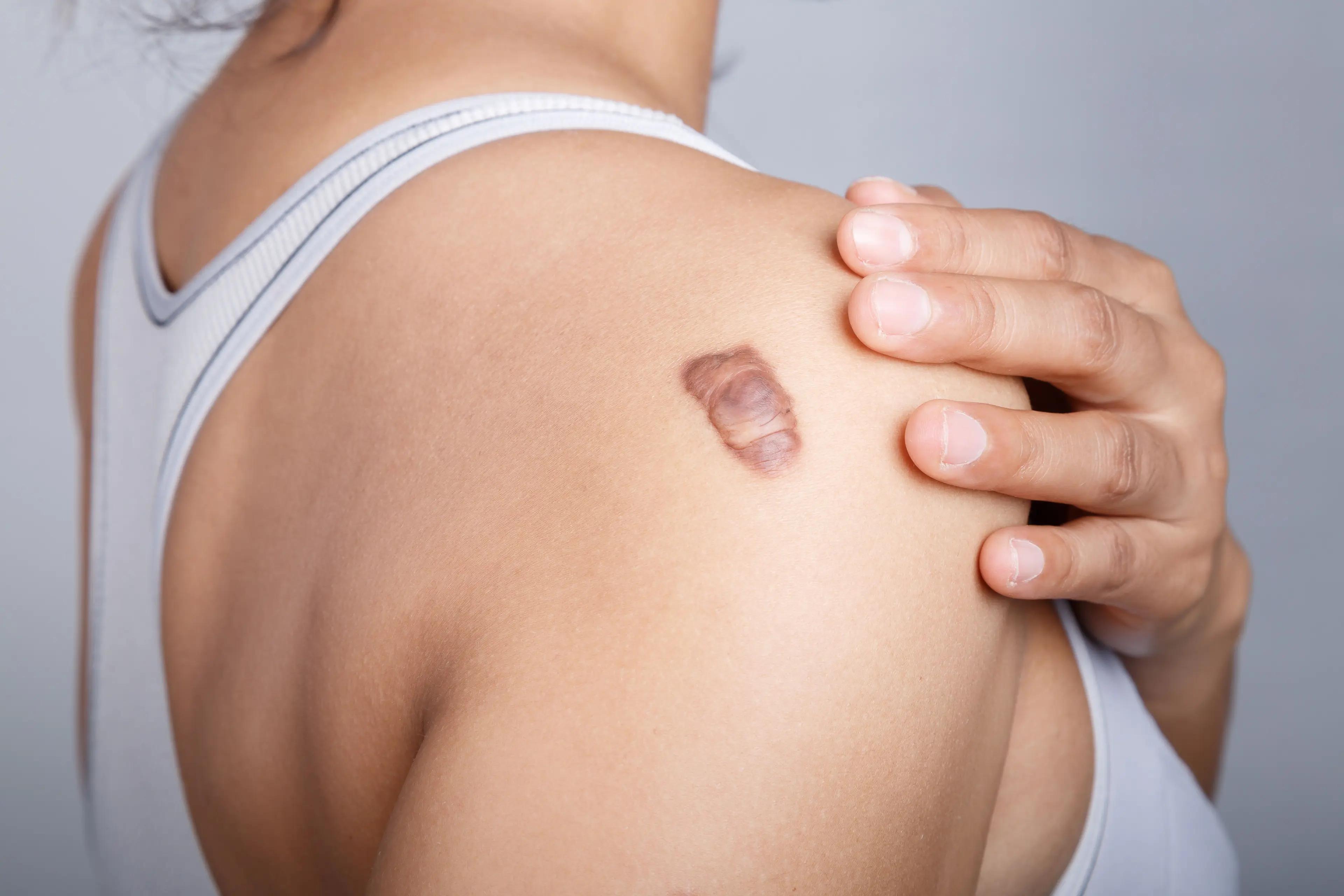
A deep learning model could accurately distinguish keloids from both benign and malignant skin lesions using nondermatoscopic clinical images, according to a study published in Diagnostics.1
The VGG16 convolutional neural network (CNN) achieved high performance metrics, including an area under the receiver operating characteristic curve of 0.996, a precision of 1.0, and an accuracy of 0.985. According to the authors, this suggests VGG16 could be a valuable tool for improving diagnostic accuracy in settings without access to dermatology specialists or dermatoscopic imaging.
As much as this proposed model performed well on test data, there were some challenges related to image quality and a relatively small amount of data that may leave out some types of lesions, warranting more research to improve the model’s generalizability.
Keloids are thick, raised scars that may appear after a skin injury, but are sometimes mistaken for malignant skin lesions since they can share similar traits such as progressive uncontrolled growth, no spontaneous regression, and high likelihoods of recurrence.2 They resemble noncancerous skin tumors caused by excessive collagen production, but unlike cancer, they do not spread to other parts of the body. As with any case, misdiagnosis can lead to delayed or improper treatment and therefore harmful outcomes.
After fine-tuning, VGG16 outperformed 8 other pretrained CNNs in 2 of 3 key classification tasks: identifying 7 types of lesions as malignant or benign and recognizing keloids as a distinct third category from malignant and benign.1 The DenseNet121 model performed best when it came to the third task of distinguishing keloids from malignant lookalikes.
To come to these findings, researchers retrained these CNNs on publicly available datasets of 2298 images of common skin cancers and 274 images of keloids. They then validated the models against test data the algorithms had not seen before. These models performed especially well on images focused on skin lesions, offering a realistic scenario for settings where there may be limited access to imaging equipment or proper expertise.
The authors tested the model on new anonymized clinical photographs to evaluate its real-world applicability. Although the VGG16 model correctly identified some test lesions, it misclassified 1 image that was cropped and blurry, highlighting the importance of image quality and framing for consistent performance and proper diagnosis.
“This is probably the result of a lack of similar data (from similar anatomical regions) used for training,” the authors said. “Hence, more image data available for training will likely improve the results. Moreover, combining the clinical features of each skin lesion with the images of these lesions has been reported to improve the performance of CNN models. We will consider such an approach in the future.”
The authors noted that training data sets were relatively small and lacked representation across all anatomical regions, and that some benign lesion images were not pathologically confirmed, which could impact reliability.
References
- Adebayo OE, Chatelain B, Trucu D, Eftimie R. Deep learning approaches for the classification of keloid images in the context of malignant and benign skin disorders. Diagnostics (Basel). 2025;15(6):710. doi:10.3390/diagnostics15060710
- Tan S, Khumalo N, Bayat A. Understanding keloid pathobiology from a quasi-neoplastic perspective: less of a scar and more of a chronic inflammatory disease with cancer-like tendencies. Front Immunol. 2019;10:1810. doi:10.3389/fimmu.2019.01810
Newsletter
Like what you’re reading? Subscribe to Dermatology Times for weekly updates on therapies, innovations, and real-world practice tips.














